Research topics
Processed pseudogenes and Retrogenes
The human genome contains approximately twenty thousand protein coding genes. During primate evolution, two distinct mechanisms mediated the creation of a similar number of protein coding genes duplications. These duplications are the source of gene fossils, gene reservoirs, and many non-coding copies, that are also called pseudogenes.
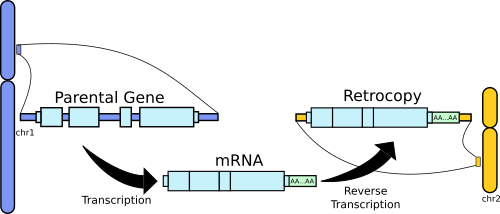
 The main mechanism creating pseudogenes is called retrotransposition. It uses mRNA templates and the reverse transcriptase from LINE-1 elements to create intron-less copies of protein coding genes at random positions of a host genome. Retrotransposition of protein coding genes transcripts was active in ancestral germinative cells during primate evolution, creating a number of retrocopies (or processed pseudogenes) that are shared between humans and other primates. The retrotransposition of protein coding transcripts is still active in humans, creating many intact protein coding genes copies that also happen to be population specific.
The main mechanism creating pseudogenes is called retrotransposition. It uses mRNA templates and the reverse transcriptase from LINE-1 elements to create intron-less copies of protein coding genes at random positions of a host genome. Retrotransposition of protein coding genes transcripts was active in ancestral germinative cells during primate evolution, creating a number of retrocopies (or processed pseudogenes) that are shared between humans and other primates. The retrotransposition of protein coding transcripts is still active in humans, creating many intact protein coding genes copies that also happen to be population specific.
How can we reliably detect these copies in complex genomes? What is the biological significance of these copies? And what is the phenotypic impact of these copies in individuals, populations, and species perspective are some of the questions I’m interested.

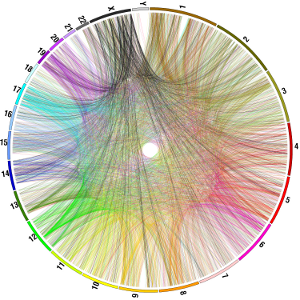
How can we reliably detect these copies in complex genomes? What is the biological significance of these copies? And what is the phenotypic impact of these copies in individuals, populations, and species perspective are some of the questions I’m interested.

Transposable Elements
Forty percent of the human genome is derived from the mobilization of Transposable Elements (TEs). The most ancient transposable elements in the human genome are DNA transposons, sequences that mobilize themselves in the genome trough a mechanism analogous to cutting and pasting. Due to their non-duplicative nature, DNA transposons are believed to be inactive and, therefore, are fossils of ancient mobilizations in our ancestral genomes.
On other hand, retrotransposons, repetitive sequences that rely on RNA-based mobilization mechanism - analogous to copying and pasting; are active in the human genome and, similar to processed pseudogenes, are polymorphic in the human lineage. LINE-1 (Long Interspersed Nuclear Element) is the most active retrotransposon in the human lineage and is believed to be one of the major mechanisms creating variability across individuals. However, due to their high copy number nature, very little is known about their activity in somatic tissue or in cancer. I'm interested in developping new methods to leverage high-throughput sequencing datasets to address their activity in the human genome.
On other hand, retrotransposons, repetitive sequences that rely on RNA-based mobilization mechanism - analogous to copying and pasting; are active in the human genome and, similar to processed pseudogenes, are polymorphic in the human lineage. LINE-1 (Long Interspersed Nuclear Element) is the most active retrotransposon in the human lineage and is believed to be one of the major mechanisms creating variability across individuals. However, due to their high copy number nature, very little is known about their activity in somatic tissue or in cancer. I'm interested in developping new methods to leverage high-throughput sequencing datasets to address their activity in the human genome.